Call:
lm(formula = Mass ~ Height, data = elephants)
Residuals:
Min 1Q Median 3Q Max
-1375.40 -193.73 -32.32 153.63 1457.10
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3553.9303 111.5451 -31.86 <2e-16 ***
Height 27.1575 0.4885 55.59 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 305.3 on 1468 degrees of freedom
Multiple R-squared: 0.6779, Adjusted R-squared: 0.6777
F-statistic: 3090 on 1 and 1468 DF, p-value: < 2.2e-16Math 430: Lecture 4b
Simple Linear Regression for Inference
Understanding the variation in our estimates
Theoretical population regression line
We said our theoretical model is \[Y_i = \beta_0 + \beta_1 X_i + \epsilon_i.\]
It is theoretical because we are saying if we had all of the data for our population, a census instead of a sample, it would be the best linear approximation for the relationship between \(X\) and \(Y\).
Realistically we will almost never have a census, so \(\beta_0\) and \(\beta_1\) are unknown in practice.
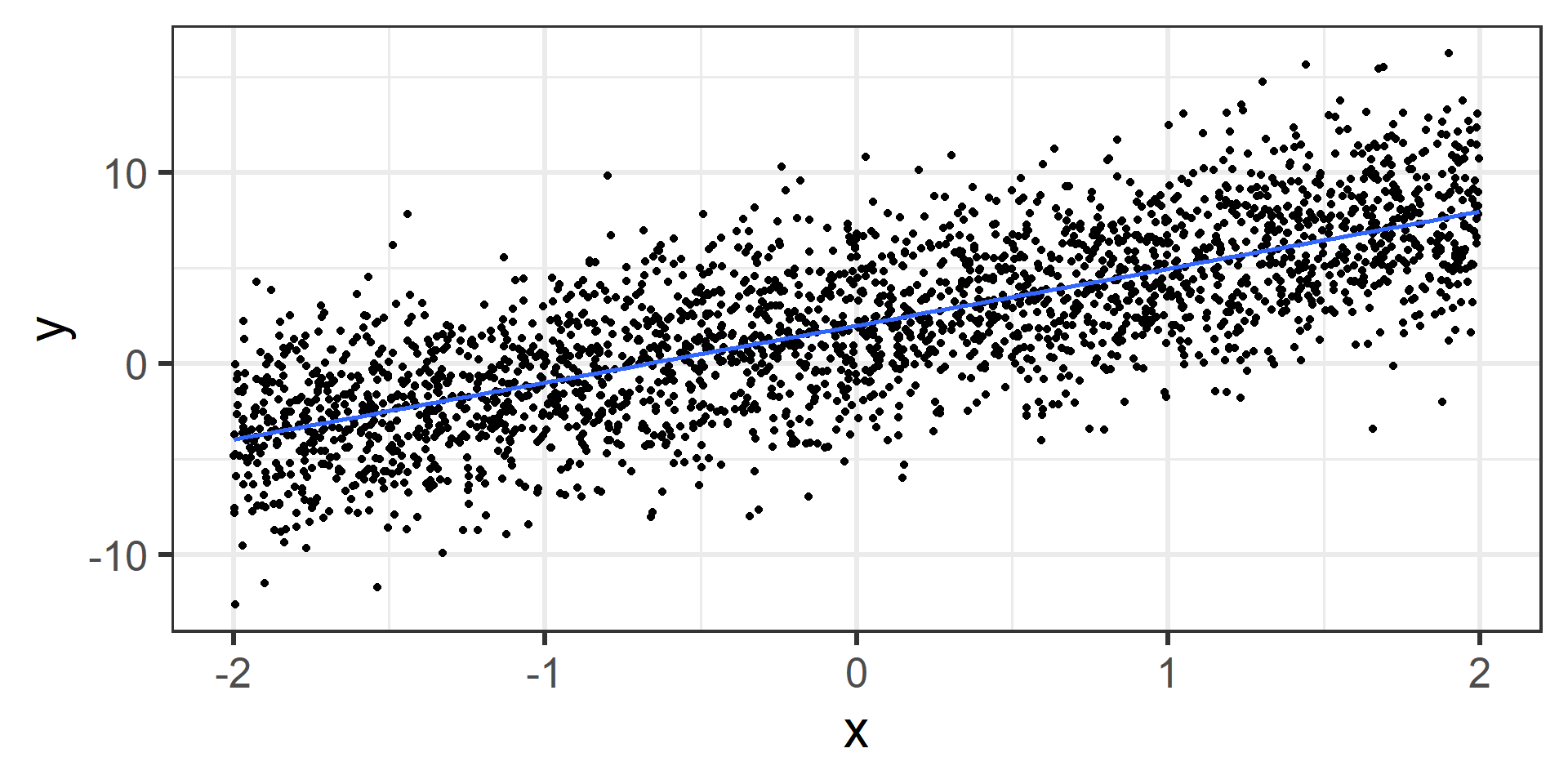
Simulation: population data
I simulated data for a population of 2500 individuals.
- randomly generate \(x\)’s (doesn’t matter how).
- randomly generate \(\epsilon\)’s where \(\epsilon_i \overset{iid}{\sim} N(0, 3^2)\).
- calculate \(y\)’s with \(y_i = 3 + 2 x_i + \epsilon_i\).
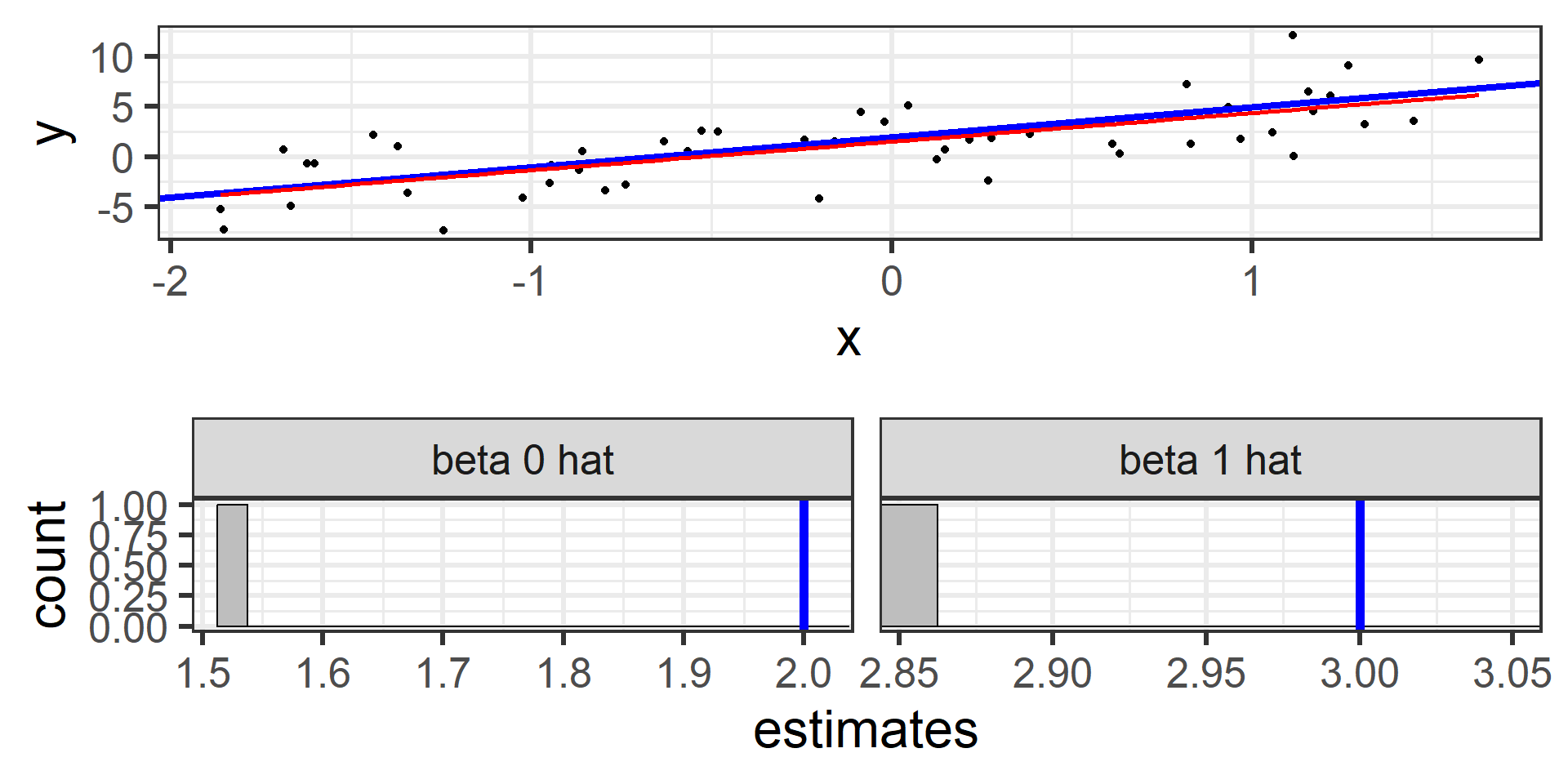
Simulation: simple random sample 1
Next I randomly sampled 50 individuals and calculate the least squares estimates \(\hat{\beta}_0\) and \(\hat{\beta}_1\)
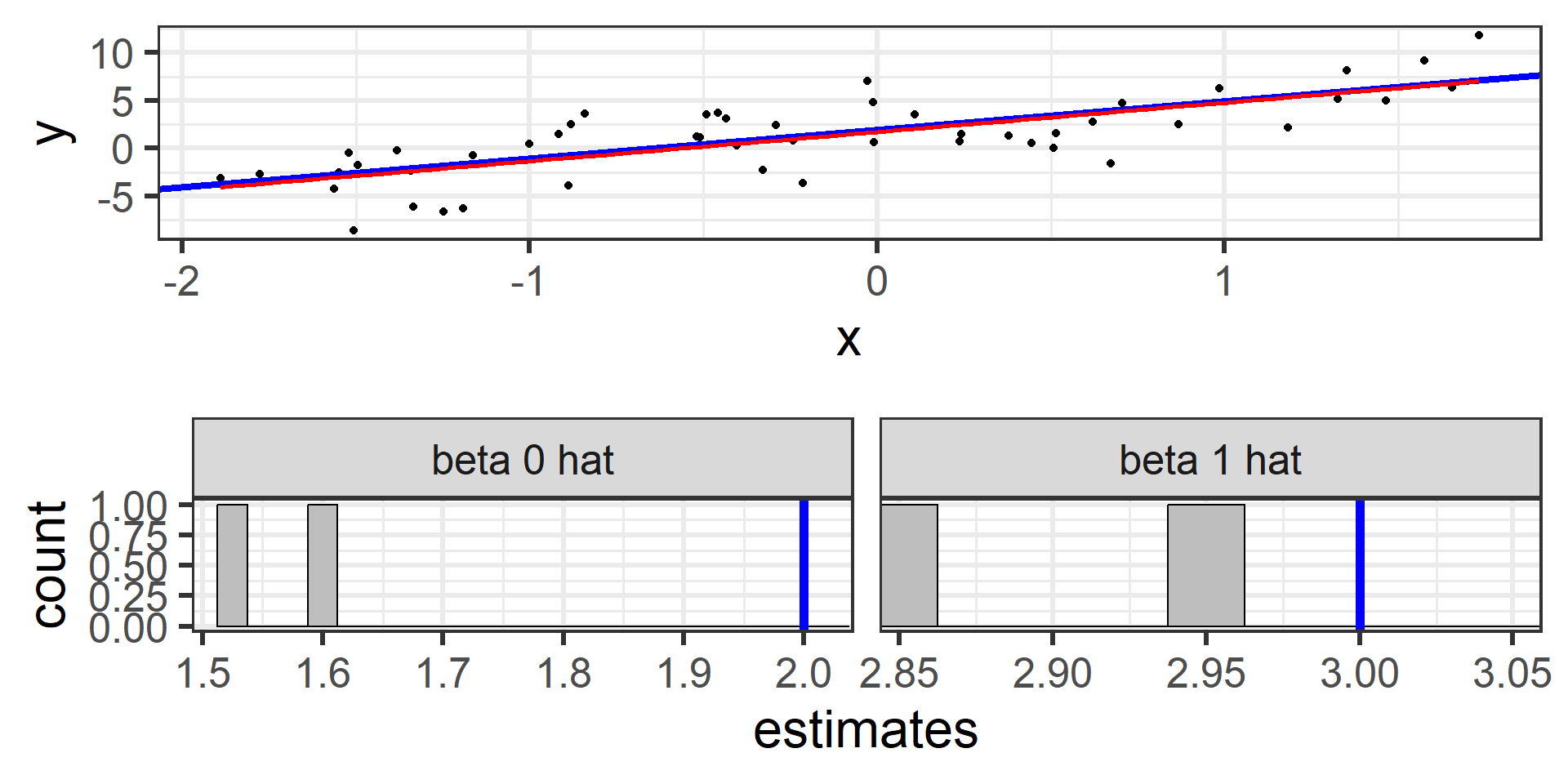
Simulation: simple random sample 2
I can do that again with another random sample of 50.
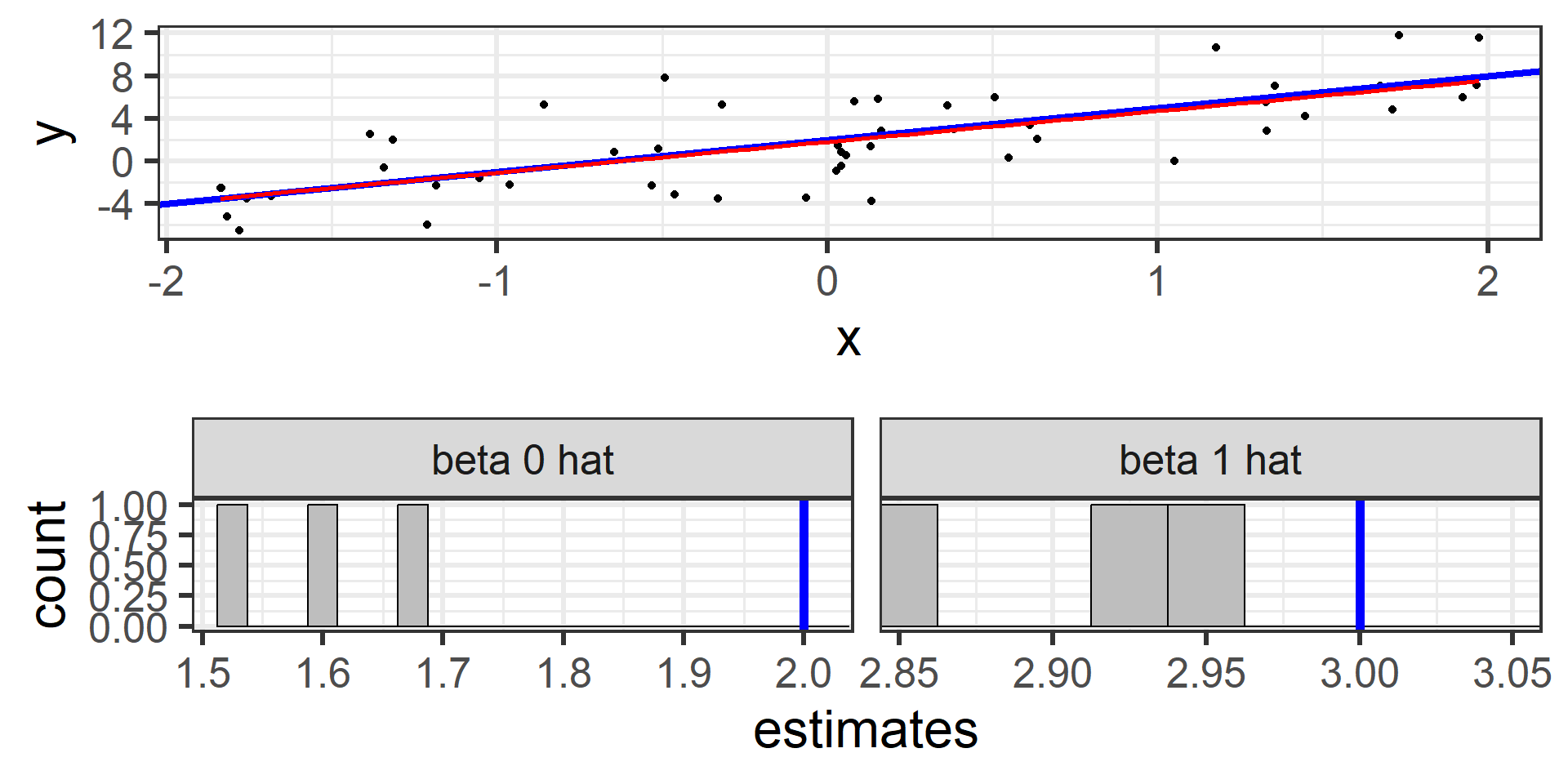
Simulation: simple random sample 3
And again with a third random sample of 50
Simulation: simple random sample 4
And again with a fourth random sample of 50
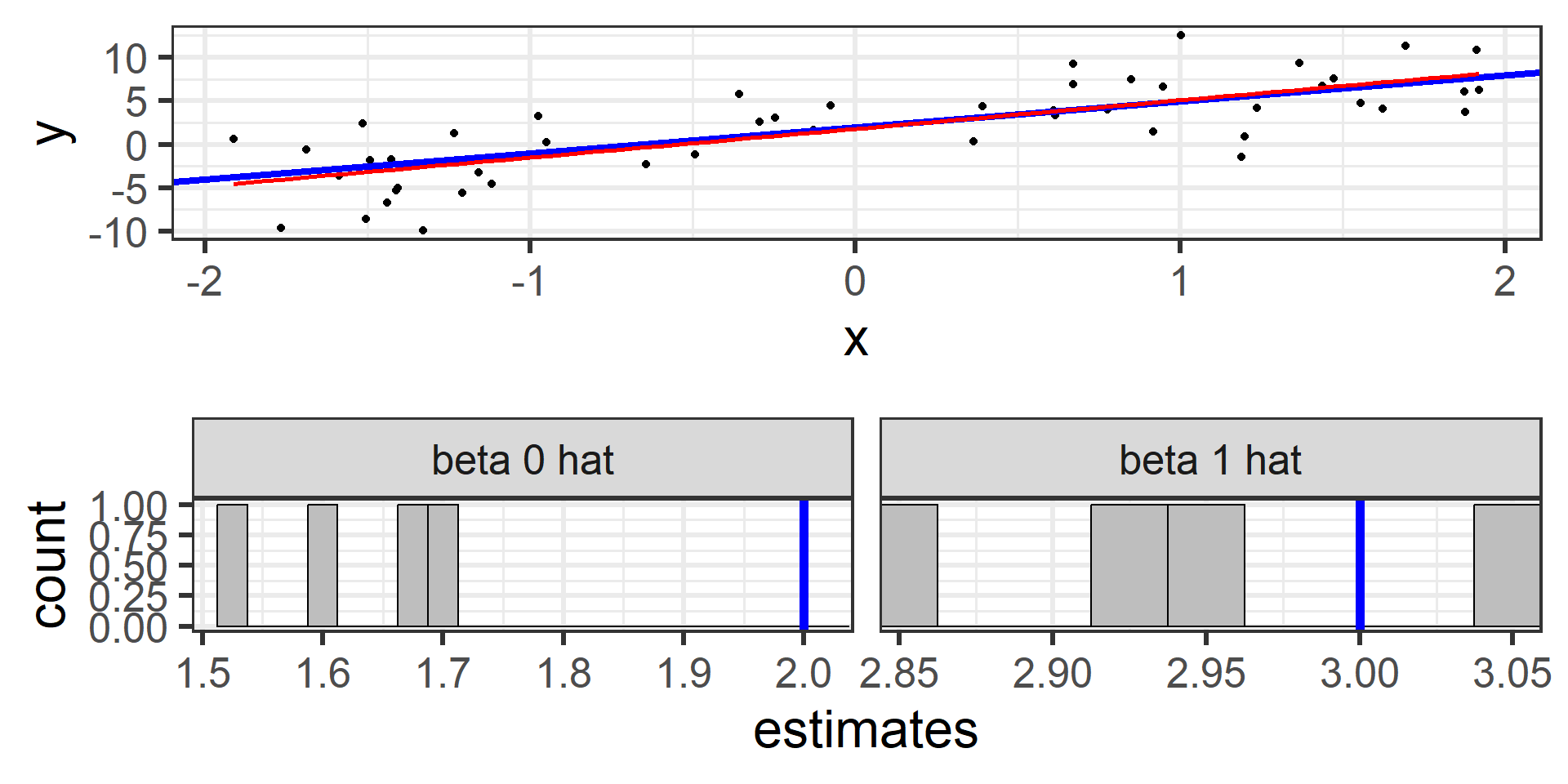
Simulation: simple random sample 5
And again with a fifth random sample of 50
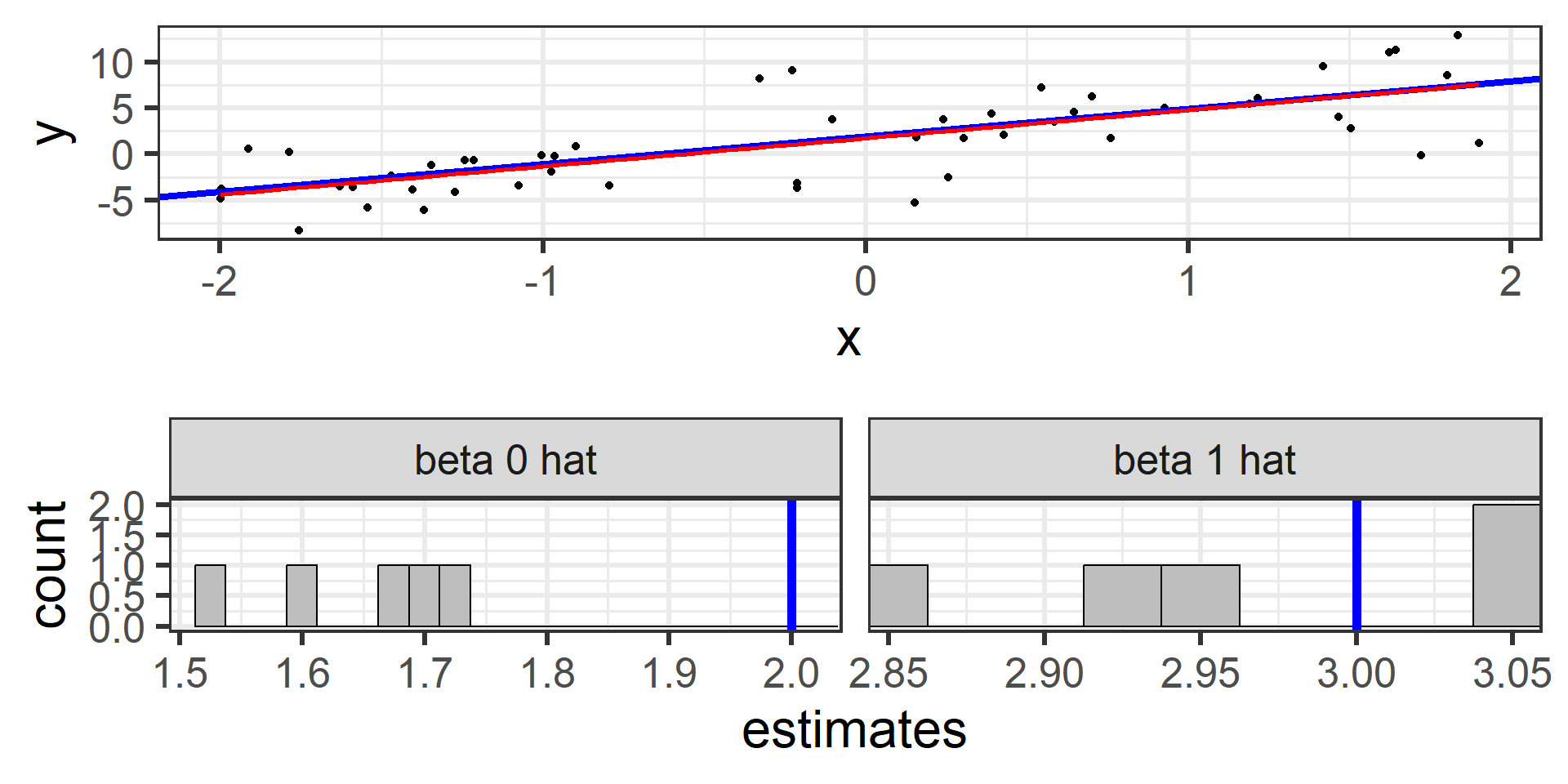
Simulation: simple random sample 50
And again 50 times!
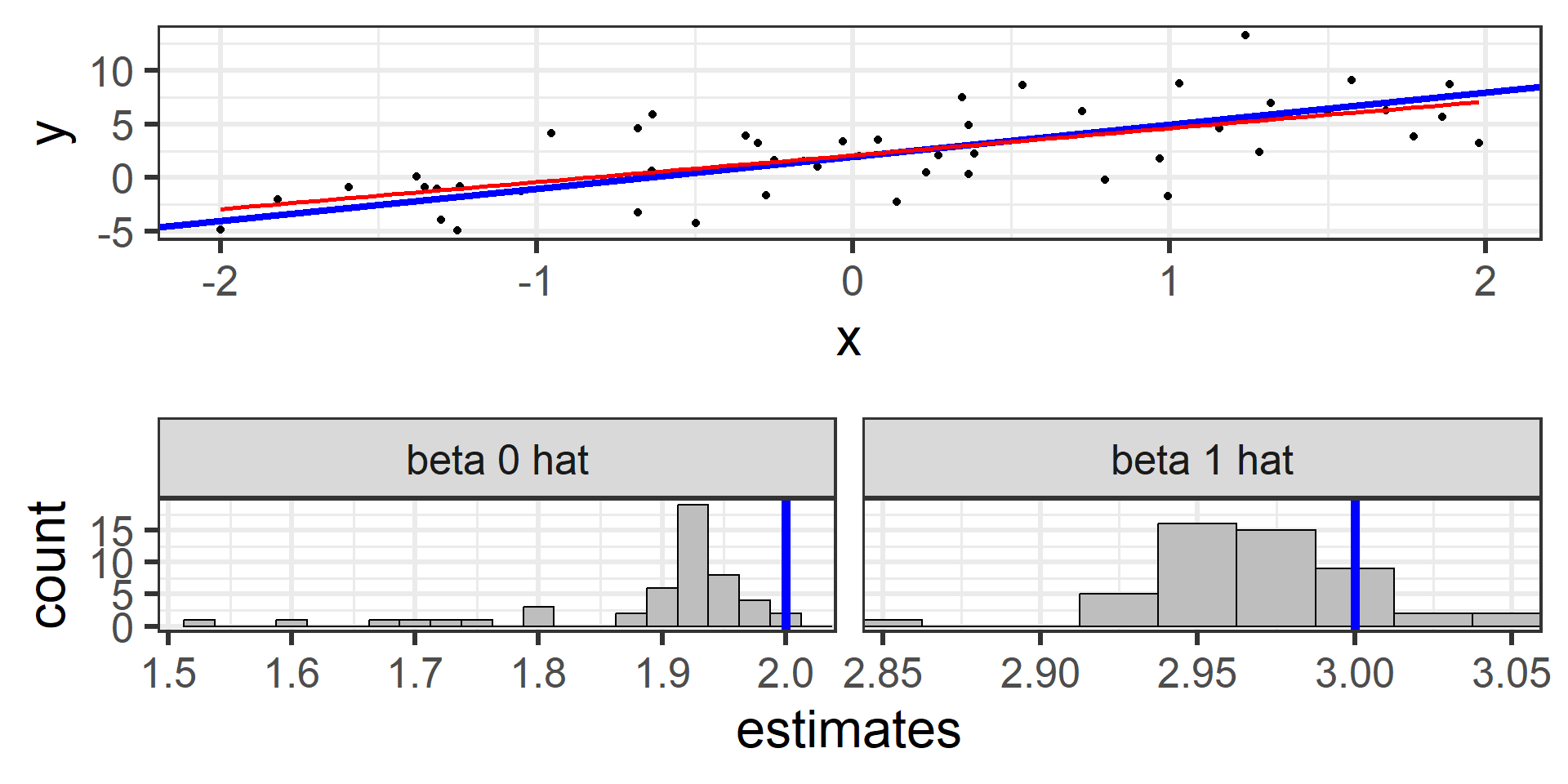
Simulation: simple random sample 100
100 times!
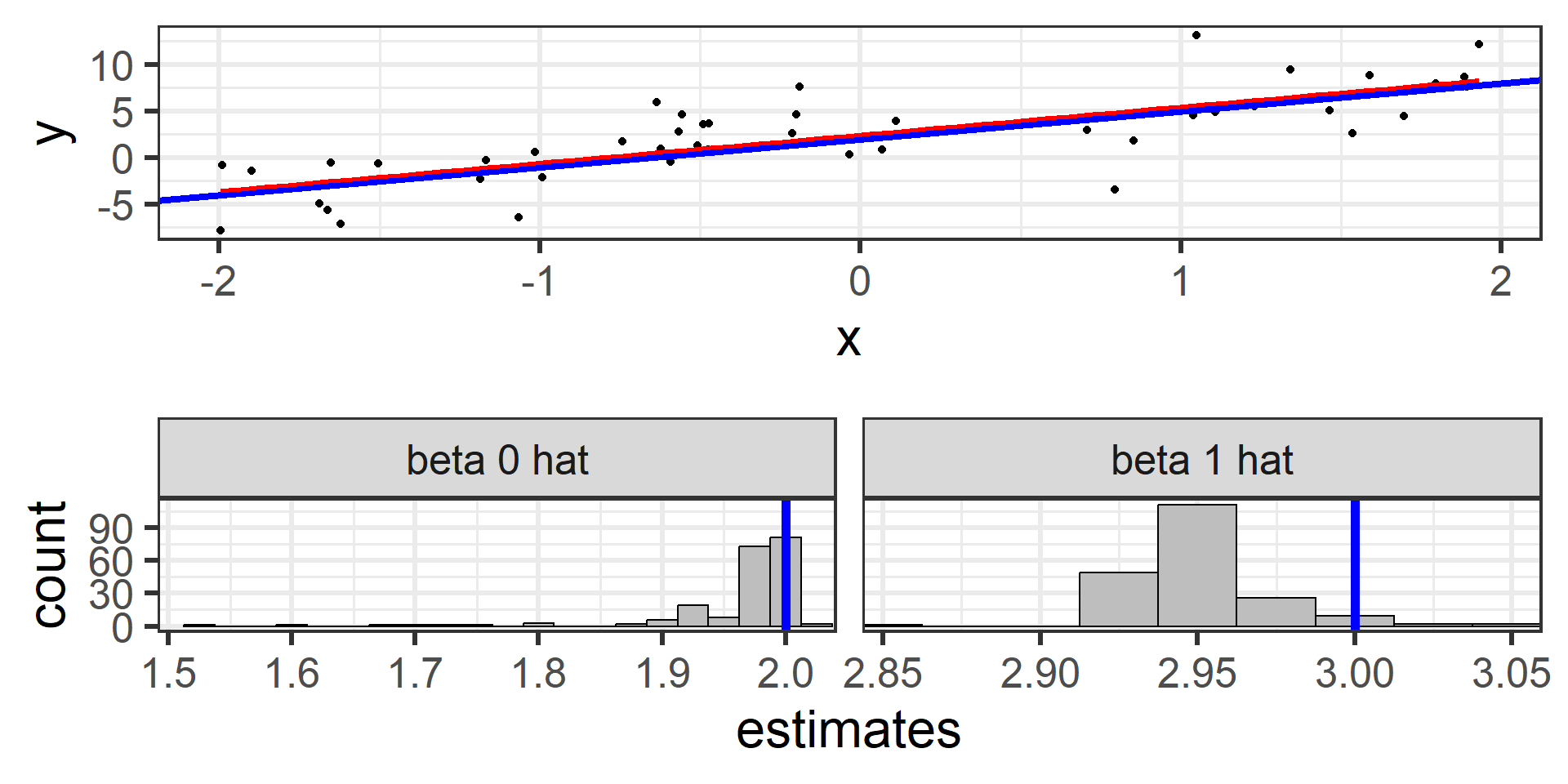
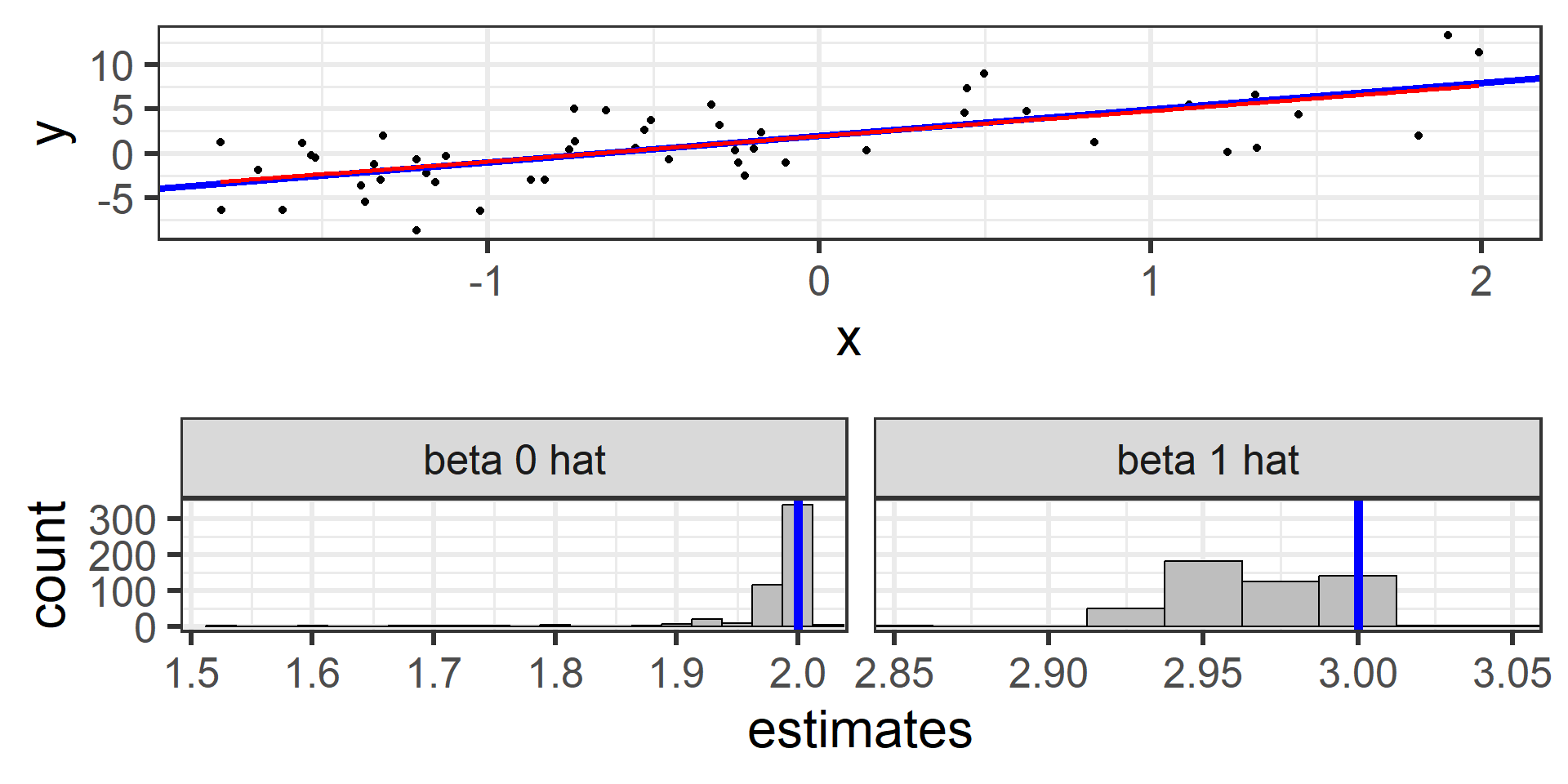
Simulation: simple random sample 500
500 times!
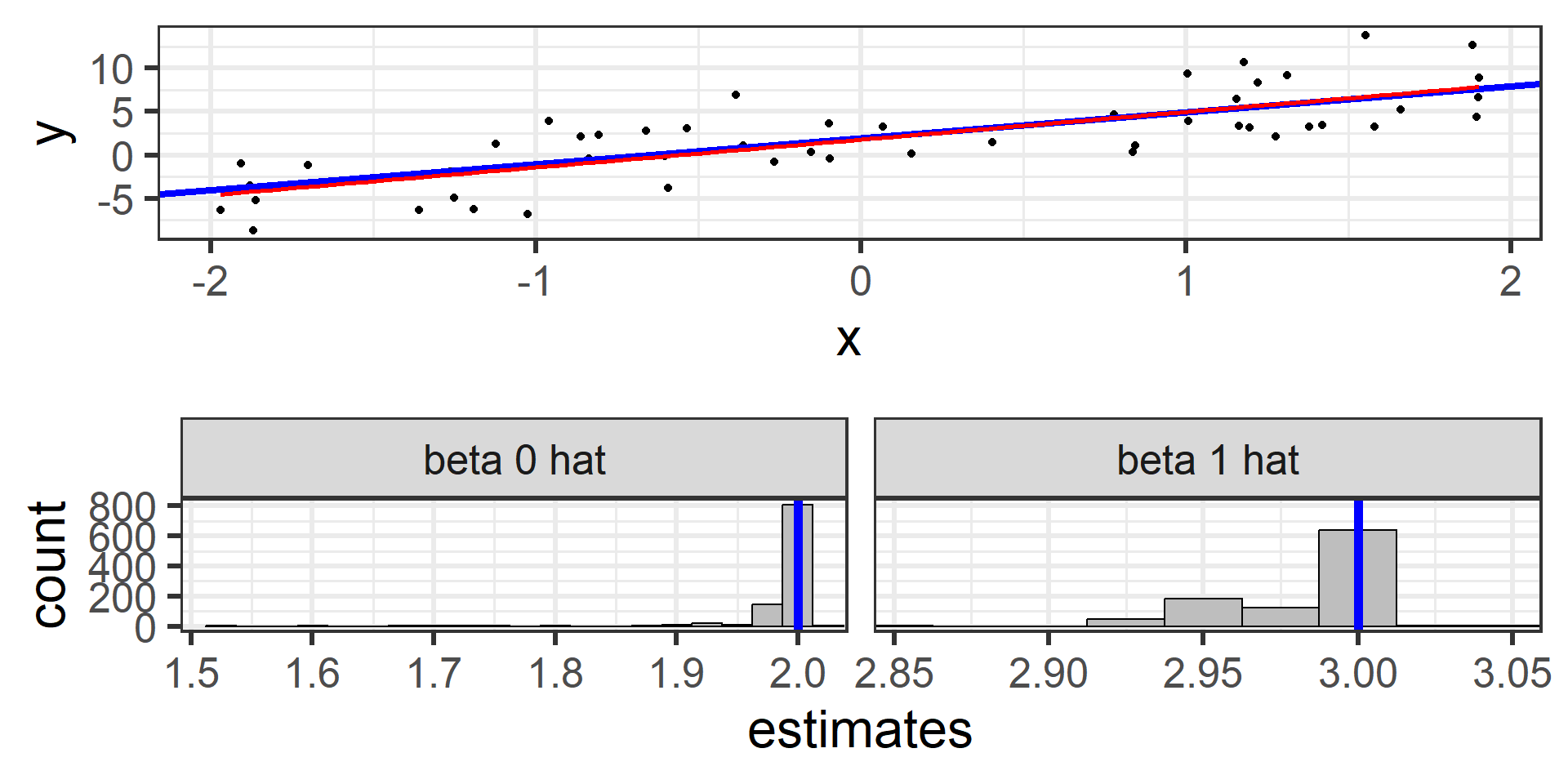
Simulation: simple random sample 1000
Even 1000 times!
Unbiased estimators
What we just saw was the sampling distribution of \(\hat{\beta}_0\) and \(\hat{\beta}_1\).
The least squares estimators \(\hat{\beta}_0\) and \(\hat{\beta}_1\) from many random samples will tend towards the true parameters \(\beta_0\) and \(\beta_1\).
\(\hat{\beta}_0\) and \(\hat{\beta}_1\) are unbiased estimators for \(\beta_0\) and \(\beta_1\)! \(E[\hat{\beta}_0] = \beta_0\) and \(E[\hat{\beta}_1] = \beta_1\)
In practice we typically only have one sample and the estimates from a single sample may be fairly different from the true parameters by chance.
This is why we should use uncertainty estimates when using our estimators!
Standard error of least squares estimates
\[SE(\hat{\beta}_0) = \sqrt{\sigma^2 \left(\frac{1}{n} + \frac{\bar{x}^2}{\sum_{i=1}^{n} (x_i - \bar{x})^2}\right)}\] \[SE(\hat{\beta}_1) = \sqrt{\frac{\sigma^2}{\sum_{i=1}^{n} (x_i - \bar{x})^2}}\]
Do you notice an issue with calculating \(SE(\hat{\beta}_0)\) and \(SE(\hat{\beta}_1)\)?
In practice
We use Residual Standard Error (RSE)
\[RSE = \hat{\sigma} = \sqrt{RSS / (n - 2),}\] since \(E[\hat{\sigma}^2] = \sigma^2\)
Technically what we calculate are estimates of the uncertainty estimators: \(\hat{SE(\hat{\beta}_0)}\) and \(\hat{SE(\hat{\beta}_1)}\)
SE and RSE in model output
Confidence Intervals
Confidence interval formula
We can use our point estimates \(\hat{\beta}_0\) and \(\hat{\beta}_1\) along with our standard error estimates \(SE(\hat{\beta}_0)\) and \(SE(\hat{\beta}_1)\) to provide confidence intervals
\[\hat{\beta}_0 \pm t^* SE(\hat{\beta}_0)\]
\[\hat{\beta}_1 \pm t^* SE(\hat{\beta}_1)\]
What is \(t^*\) ?
Technically \(t^* = t_{(1 - CL) / 2, df = n - 2}\).
For this class we won’t focus on the exact calculation of \(t^*\), we will just provide an understanding.
It essentially uses the confidence level (CL) to adjust the width of our confidence interval.
Higher confidence level results in wider confidence intervals
The textbook points out \(t^* \approx 2\) for 95% confidence intervals with large sample size \(n\)
Computing confidence intervals in practice
The confint() function will computer 95% confidence intervals by default
2.5 % 97.5 %
(Intercept) -3772.73505 -3335.12552
Height 26.19922 28.11584If we want a different confidence level we have to specify that
Interpreting confidence intervals
Confidence level \(\neq\) probability
The confidence interval is the long-term proportion of confidence intervals that will capture the true parameter (either \(\beta_0\) or \(\beta_1\))
For elephants 0 cm tall, the mass is expected to be between -3696 kg and -3410 kg, with 95% confidence.
A 1 cm increase in height of elephants is associated with an higher mass of between 26.5 kg to 27.7 kg, with 95% confidence.
Hypothesis testing
Hypotheses
\(H_0\): There is no linear relationship between \(Y\) and \(X\).
\(H_1\): There is a linear relationship between \(Y\) and \(X\).
Or equivalently:
\(H_0\): \(\beta_1 = 0\)
\(H_1\): \(\beta_1 \neq 0\)
For hypotheses tests we assume the null is true (\(H_0\)), and see if we can find evidence against it (i.e., low p-value), in support of the alternative hypothesis (\(H_1\))
Test statistic
\(t = \frac{\hat{\beta}_1 - 0}{SE(\hat{\beta}_1)}\)
Call:
lm(formula = Mass ~ Height, data = elephants)
Residuals:
Min 1Q Median 3Q Max
-1375.40 -193.73 -32.32 153.63 1457.10
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3553.9303 111.5451 -31.86 <2e-16 ***
Height 27.1575 0.4885 55.59 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 305.3 on 1468 degrees of freedom
Multiple R-squared: 0.6779, Adjusted R-squared: 0.6777
F-statistic: 3090 on 1 and 1468 DF, p-value: < 2.2e-16p-value
The p-value is <2e-16, which is essentially zero.
The p-value is the probability of a test statistics as or more extreme as what we observed, if the null hypothesis were true.
So it is extremely improbable to observe what we observed if there was no linear relationship between mass and height of elephants (p-value <2e-16).
This means we found evidence of a linear association!
Correspondence between hypothesis tests and p-value
If a confidence interval contains zero, the p-value will be less than 1 - Confidence Level